The SciSpacy challenge from AllenAI supplies a language mannequin educated on biomedical textual content, which can be utilized for Named Entity Recognition (NER) of biomedical entities utilizing the usual SpaCy API. In contrast to the entities discovered utilizing SpaCy’s language fashions (at the very least the English one), the place entities have sorts equivalent to PER, GEO, ORG, and so forth., SciSpacy entities have the one sort ENTITY. With a purpose to additional classify them, SciSpacy supplies Entity Linking (NEL) performance by way of its integration with numerous ontology suppliers, such because the Unified Medical Language System (UMLS), Medical Topic Headings (MeSH), RxNorm, Gene Ontology (GO), and Human Phenotype Ontology (HPO).
The NER and NEL processes are decoupled. The NER course of finds candidate entity spans, and these spans are matched towards the respective ontologies, which can end result within the span matching zero or extra ontology entries. All candidate span is then matched to all of the matched entities.
On this publish, I’ll describe a technique to disambiguate the linked entities. Primarily based on restricted testing, this chooses the right idea about 73% of the time.
The technique relies on the instinct that an ambiguously linked entity span is extra prone to resolve to an idea that’s carefully associated to ideas for the opposite non-ambiguously linked entity spans within the sentence. In different phrases, one of the best goal label to decide on for an ambiguous entity is the one that’s semantically closest to the labels of different entities within the sentence. Or much more succintly, and with apologies to John Firth, an entity is thought by the corporate it retains.
The truth that viral antigens couldn’t be demonstrated with the used staining isn’t the results of antibodies current within the cat that already sure to those antigens and hinder binding of different antibodies.
The NEL step will try and match these spans towards the UMLS ontology. Outcomes for the matching are proven under. As famous earlier, every UMLS idea maps to a number of sematic sorts, and these are proven right here as effectively.
| Entity-ID | Entity Span | Idea-ID | Idea Main Identify | Semantic Kind Code | Semantic Kind Identify |
|---|---|---|---|---|---|
| 1 | staining | C0487602 | Staining technique | T059 | Laboratory Process |
| 2 | antibodies | C0003241 | Antibodies | T116 | Amino Acid, Peptide, or Protein |
| T129 | Immunologic Issue | ||||
| 3 | cat | C0007450 | Felis catus | T015 | Mammal |
| C0008169 | Chloramphenicol O-Acetyltransferase | T116 | Amino Acid, Peptide, or Protein | ||
| T126 | Enzyme | ||||
| C0325089 | Household Felidae | T015 | Mammal | ||
| C1366498 | Chloramphenicol Acetyl Transferase Gene | T028 | Gene or Genome | ||
| 4 | antigens | C0003320 | Antigens | T129 | Immunologic Issue |
| 5 | binding | C1145667 | Binding motion | T052 | Exercise |
| C1167622 | Binding (Molecular Operate) | T044 | Molecular Operate | ||
| 6 | antibodies | C0003241 | Antibodies | T116 | Amino Acid, Peptide, or Protein |
| T129 | Immunologic Issue |
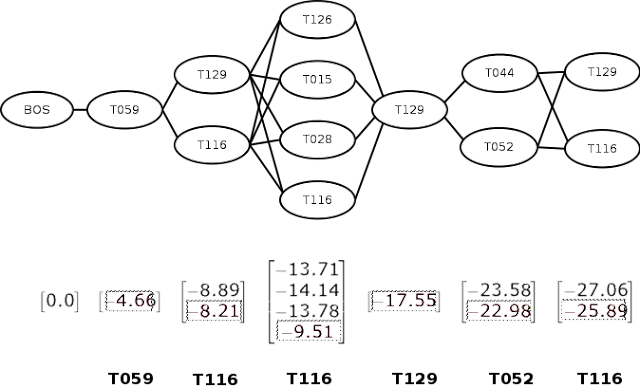
The sequence of entity spans, every mapped to a number of semantic sort codes might be represented by a graph of semantic sort nodes as proven under. Right here, every vertical grouping corresponds to an entity place. The BOS node is a particular node representing the start of the sequence. Primarily based on our instinct above, entity disambiguation is now only a matter of discovering the almost certainly path by way of the graph.
The Viterbi algorithm consists of two phases — ahead and backward. Within the ahead part, we transfer left to proper, computing the log-probability of every transition at every step, as proven by the vectors under every place within the determine. When computing the transition from a number of nodes to a single node (such because the one from [T129, T116] to [T126], we compute for each paths and select the utmost worth.
Within the backward part, we transfer from proper to left, selecting the utmost chance node at every step. That is proven within the determine as boxed entries. We are able to then lookup the suitable semantic sort and return the almost certainly sequence of semantic sorts (proven in daring within the backside of the determine).
Nevertheless, our goal is to return disambiguated idea linkages for entities. Given a disambiguated semantic sort and a number of potentialities indicated by SciSpacy’s linking course of, we use the emission chances to decide on the almost certainly idea to use on the place. The end result for our instance is proven within the desk under.
| Entity-ID | Entity Span | Idea-ID | Idea Main Identify | Semantic Kind Code | Semantic Kind Identify | Right? |
|---|---|---|---|---|---|---|
| 1 | staining | C0487602 | Staining technique | T059 | Laboratory Process | N/A* |
| 2 | antibodies | C0003241 | Antibodies | T116 | Amino Acid, Peptide, or Protein | Sure |
| 3 | cat | C0008169 | Chloramphenicol O-Acetyltransferase | T116 | Amino Acid, Peptide, or Protein | No |
| 4 | antigens | C0003320 | Antigens | T129 | Immunologic Issue | N/A* |
| 5 | binding | C1145667 | Binding motion | T052 | Exercise | Sure |
| 6 | antibodies | C0003241 | Antibodies | T116 | Amino Acid, Peptide, or Protein | Sure |
(N/A: non-ambiguous mappings)
- Code: This github gist incorporates code that illustrates NER + NEL on an enter sentence utilizing SciSpacy and its UMLS integration, after which applies my adaptation of the Viterbi technique (as described on this publish) to disambiguate ambiguous entity linkages.
- Information: I’ve additionally offered the transition and emission matrices, and their related lookup tables, for comfort, as these might be time consuming to generate from scratch from the CORD-19 dataset.
As all the time, I respect your suggestions. Please let me know when you discover flaws with my strategy, and/or you understand of a greater strategy for entity disambiguation